Dataset Preparation¶
1. Introduction¶
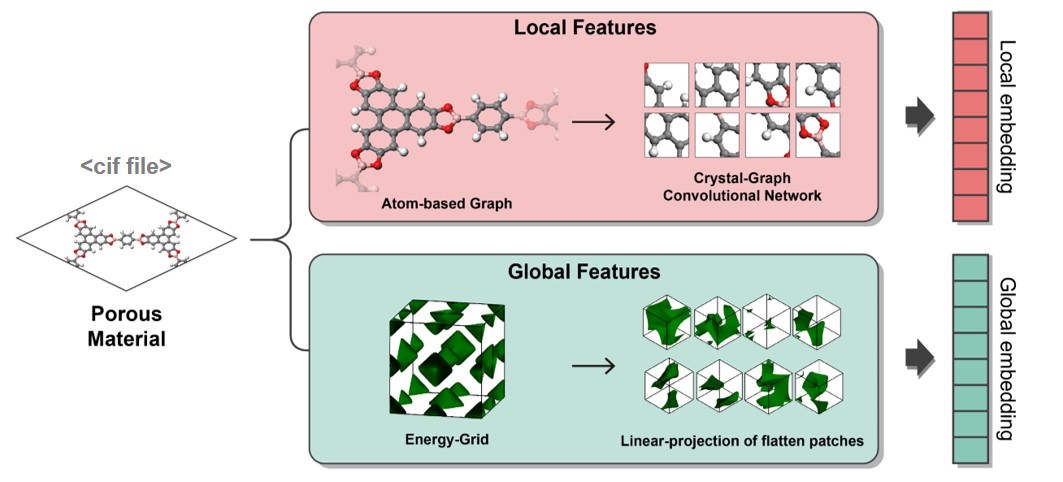
MOFTransformer takes both atom-wise graph embeddings and energy-grid embeddings to capture local and global features,respectively. Both of embeddings are generated from the cif files of MOFs.
(1) atom-wise graph embeddings
Tha atom-wise graph embeddings are taken from the modified CGCNN by removing pooling layer and adding opologically unique atom selection.
(2) energy-grid embeddings
The 3D energy grids are calculated using GRIDAY with the united atom model of methane molecule using UFF.
2.Generate custom dataset¶
To generate inputs for MOFTransformer from CIF files, use the moftransformer/utils/prepare_data file to generate atom-wise graph embeddings and energy-grid embeddings. You will need to prepare CIF files and raw_{downstream}.json files in a root_cifs directory.
root_cif: A directory that contains.cifand.jsonfile.downstream: name of user-specific downstream task (e.g. band_gap, gas_uptake, etc).
Example for root_cif¶
The example of root_cifs directory is as follows.
root_cifs # root for cif files
├── [cif_id].cif
├── [cif_id].cif
├── ...
└── raw_{downstream}.json
The example of raw_{downstream}.json files is as follows.
{
cif_id : property (float or int),
...
}
Run prepare_data function¶
If there is a json files named raw_{downstream}.json in root_cifs directory, then it will be randomly splitted to train, val and test. (Default = 8:1:1).
from moftransformer.utils import prepare_data
# single task
prepare_data(root_cifs, root_dataset, downstream="example")
# multiple tasks (contain several json files in root_cif)
prepare_data(root_cifs, root_dataset, downstream=["example1", "example2", ...])
prepare_data will generate the atom-wise graph embeddings and energy-grid embeddings in root_dataset
directory.
root_dataset # root for generated inputs
├── train
│ ├── [cif_id].graphdata # graphdata
│ ├── [cif_id].grid # energy grid information
│ ├── [cif_id].griddata16 # grid data
│ ├── [cif_id].cif # primitive cif
│ └── ...
├── val
│ ├── [cif_id].graphdata # graphdata
│ ├── [cif_id].grid # energy grid information
│ ├── [cif_id].griddata16 # grid data
│ ├── [cif_id].cif # primitive cif
│ └── ...
├── test
│ ├── [cif_id].graphdata # graphdata
│ ├── [cif_id].grid # energy grid information
│ ├── [cif_id].griddata16 # grid data
│ ├── [cif_id].cif # primitive cif
│ └── ...
├── train_{downstream}.json
├── val_{downstream}.json
└── test_{downstream}.json
3. Dataset for public database (CoREMOF, QMOF).¶
we’ve provided the dataset of atom-wise graph embedding and energy-grid embedding for the CoREMOF and the QMOF database in our figshare database.
Or, you can download using command line or python (refer to installation)